1 Introduction
In my last post, I showed how to do binary classification using the Keras deep learning library. Now I would like to show how to make multi-class classifications.
For this publication the dataset bird from the statistic platform “Kaggle” was used. You can download it from my “GitHub Repository”.
2 Loading the libraries
import pandas as pd
import numpy as np
import os
import shutil
import pickle as pk
import matplotlib.pyplot as plt
from sklearn.preprocessing import OneHotEncoder
from sklearn.model_selection import train_test_split
from keras import models
from keras import layers
from keras.callbacks import EarlyStopping, ModelCheckpoint
from keras.models import load_model3 Loading the data
df = pd.read_csv('bird.csv').dropna()
print()
print('Shape of dataframe:')
print(str(df.shape))
print()
print('Head of dataframe:')
df.head()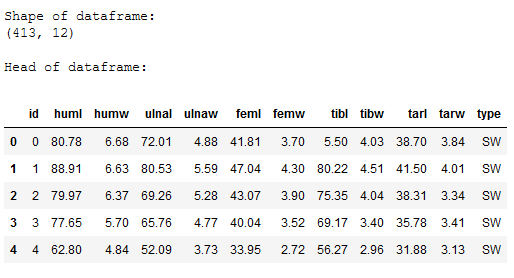
Description of predictors:
- Length and Diameter of Humerus
- Length and Diameter of Ulna
- Length and Diameter of Femur
- Length and Diameter of Tibiotarsus
- Length and Diameter of Tarsometatarsus
df['type'].value_counts()
Description of the target variable:
- SW: Swimming Birds
- W: Wading Birds
- T: Terrestrial Birds
- R: Raptors
- P: Scansorial Birds
- SO: Singing Birds
4 Data pre-processing
4.2 Encoding
Last time (Artificial Neural Network for binary Classification) we used LabelEncoder for this. Since we now want to do a multi-class classification we need One-Hot Encoding.
encoder = OneHotEncoder()
encoded_Y = encoder.fit(y.values.reshape(-1,1))
encoded_Y = encoded_Y.transform(y.values.reshape(-1,1)).toarray()
encoded_Y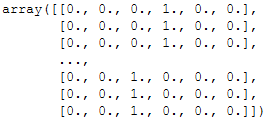
4.3 Train-Validation-Test Split
In the following, I will randomly assign 70% of the data to the training part and 15% each to the validation and test part.
train_ratio = 0.70
validation_ratio = 0.15
test_ratio = 0.15
# Generate TrainX and TrainY
trainX, testX, trainY, testY = train_test_split(x, encoded_Y, test_size= 1 - train_ratio)
# Genearate ValX, TestX, ValY and TestY
valX, testX, valY, testY = train_test_split(testX, testY, test_size=test_ratio/(test_ratio + validation_ratio))print(trainX.shape)
print(valX.shape)
print(testX.shape)
4.4 Check if all classes are included in every split part
Since this is a very small dataset with 413 observations and the least represented class contains only 23 observations, I advise at this point to check whether all classes are included in the variables created by the train validation test split.
re_transformed_array_trainY = encoder.inverse_transform(trainY)
unique_elements, counts_elements = np.unique(re_transformed_array_trainY, return_counts=True)
unique_elements_and_counts_trainY = pd.DataFrame(np.asarray((unique_elements, counts_elements)).T)
unique_elements_and_counts_trainY.columns = ['unique_elements', 'count']
unique_elements_and_counts_trainY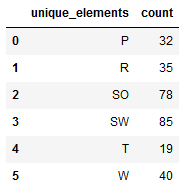
re_transformed_array_valY = encoder.inverse_transform(valY)
unique_elements, counts_elements = np.unique(re_transformed_array_valY, return_counts=True)
unique_elements_and_counts_valY = pd.DataFrame(np.asarray((unique_elements, counts_elements)).T)
unique_elements_and_counts_valY.columns = ['unique_elements', 'count']
unique_elements_and_counts_valY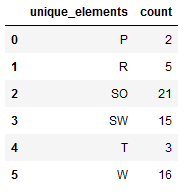
re_transformed_array_testY = encoder.inverse_transform(testY)
unique_elements, counts_elements = np.unique(re_transformed_array_testY, return_counts=True)
unique_elements_and_counts_testY = pd.DataFrame(np.asarray((unique_elements, counts_elements)).T)
unique_elements_and_counts_testY.columns = ['unique_elements', 'count']
unique_elements_and_counts_testY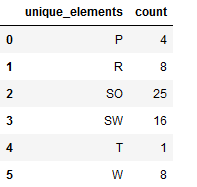
Of course, you can also use a for-loop:
y_part = [trainY, valY, testY]
for y_part in y_part:
re_transformed_array = encoder.inverse_transform(y_part)
unique_elements, counts_elements = np.unique(re_transformed_array, return_counts=True)
unique_elements_and_counts = pd.DataFrame(np.asarray((unique_elements, counts_elements)).T)
unique_elements_and_counts.columns = ['unique_elements', 'count']
print('---------------')
print(unique_elements_and_counts)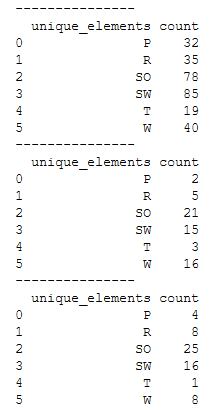
To check if all categories are contained in all three variables (trainY, valY and testY) I first store the unique elements in lists and can then compare them with a logical query.
list_trainY = unique_elements_and_counts_trainY['unique_elements'].to_list()
list_valY = unique_elements_and_counts_valY['unique_elements'].to_list()
list_testY = unique_elements_and_counts_testY['unique_elements'].to_list()
print(list_trainY)
print(list_valY)
print(list_testY)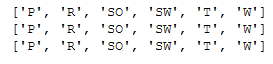
check_val = all(item in list_valY for item in list_trainY)
if check_val is True:
print('OK !')
print("The list_valY contains all elements of the list_trainY.")
else :
print()
print('No !')
print("List_valY doesn't have all elements of the list_trainY.")
check_test = all(item in list_testY for item in list_trainY)
if check_test is True:
print('OK !')
print("The list_testY contains all elements of the list_trainY.")
else :
print()
print('No !')
print("List_testY doesn't have all elements of the list_trainY.")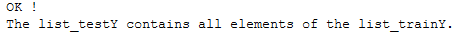
5 ANN for Multi-Class Classfication
5.2 Parameter Settings
input_shape = trainX.shape[1]
n_batch_size = 20
n_steps_per_epoch = int(trainX.shape[0] / n_batch_size)
n_validation_steps = int(valX.shape[0] / n_batch_size)
n_test_steps = int(testX.shape[0] / n_batch_size)
n_epochs = 25
num_classes = trainY.shape[1]
print('Input Shape: ' + str(input_shape))
print('Batch Size: ' + str(n_batch_size))
print()
print('Steps per Epoch: ' + str(n_steps_per_epoch))
print()
print('Validation Steps: ' + str(n_validation_steps))
print('Test Steps: ' + str(n_test_steps))
print()
print('Number of Epochs: ' + str(n_epochs))
print()
print('Number of Classes: ' + str(num_classes))
5.3 Layer Structure
Here I use the activation function ‘softmax’ in contrast to the binary classification.
model = models.Sequential()
model.add(layers.Dense(64, activation='relu', input_shape=(input_shape,)))
model.add(layers.Dense(64, activation='relu'))
model.add(layers.Dense(num_classes, activation='softmax'))model.summary()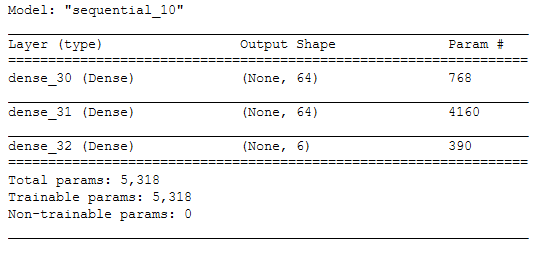
5.4 Configuring the model for training
Again, the neural network for multi-class classification differs from that for binary classification. Here the loss function ‘categorical_crossentropy’ is used.
model.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['accuracy'])5.5 Callbacks
If you want to know more about callbacks you can read about it here at Keras or also in my post about Convolutional Neural Networks.
# Prepare a directory to store all the checkpoints.
checkpoint_dir = './'+ checkpoint_no
if not os.path.exists(checkpoint_dir):
os.makedirs(checkpoint_dir)keras_callbacks = [ModelCheckpoint(filepath = checkpoint_dir + '/' + model_name,
monitor='val_loss', save_best_only=True, mode='auto')]5.6 Fitting the model
history = model.fit(trainX,
trainY,
steps_per_epoch=n_steps_per_epoch,
epochs=n_epochs,
batch_size=n_batch_size,
validation_data=(valX, valY),
validation_steps=n_validation_steps,
callbacks=[keras_callbacks])
5.7 Obtaining the best model values
hist_df = pd.DataFrame(history.history)
hist_df['epoch'] = hist_df.index + 1
cols = list(hist_df.columns)
cols = [cols[-1]] + cols[:-1]
hist_df = hist_df[cols]
hist_df.to_csv(checkpoint_no + '/' + 'history_df_' + model_name + '.csv')
hist_df.head()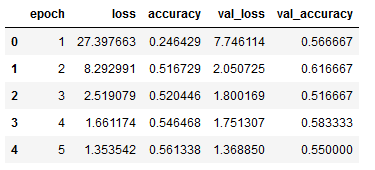
values_of_best_model = hist_df[hist_df.val_loss == hist_df.val_loss.min()]
values_of_best_model
5.8 Obtaining class assignments
Similar to the neural networks for computer vision, I also save the class assignments for later reuse.
class_assignment = dict(zip(y, encoded_Y))
df_temp = pd.DataFrame([class_assignment], columns=class_assignment.keys())
df_temp = df_temp.stack()
df_temp = pd.DataFrame(df_temp).reset_index().drop(['level_0'], axis=1)
df_temp.columns = ['Category', 'Allocated Number']
df_temp.to_csv(checkpoint_no + '/' + 'class_assignment_df_' + model_name + '.csv')
print('Class assignment:')
class_assignment
The encoder used is also saved.
pk.dump(encoder, open(checkpoint_no + '/' + 'encoder.pkl', 'wb'))5.9 Validation
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(1, len(acc) + 1)
plt.plot(epochs, acc, 'bo', label='Training acc')
plt.plot(epochs, val_acc, 'b', label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.figure()
plt.plot(epochs, loss, 'bo', label='Training loss')
plt.plot(epochs, val_loss, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.show()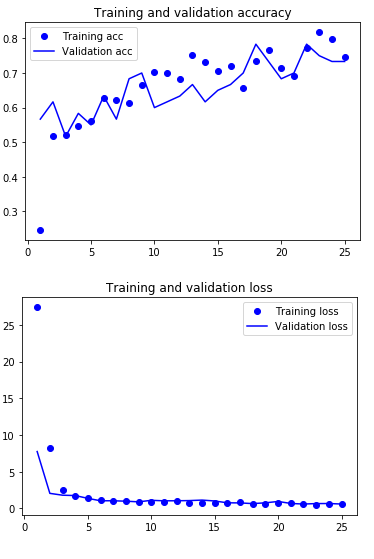
5.10 Load best model
Again, reference to the Computer Vision posts where I explained why and how I cleaned up the Model Checkpoint folders.
# Loading the automatically saved model
model_reloaded = load_model(checkpoint_no + '/' + model_name)
# Saving the best model in the correct path and format
root_directory = os.getcwd()
checkpoint_dir = os.path.join(root_directory, checkpoint_no)
model_name_temp = os.path.join(checkpoint_dir, model_name + '.h5')
model_reloaded.save(model_name_temp)
# Deletion of the automatically created folder under Model Checkpoint File.
folder_name_temp = os.path.join(checkpoint_dir, model_name)
shutil.rmtree(folder_name_temp, ignore_errors=True)best_model = load_model(model_name_temp)The overall folder structure should look like this:
5.11 Model Testing
test_loss, test_acc = best_model.evaluate(testX,
testY,
steps=n_test_steps)
print()
print('Test Accuracy:', test_acc)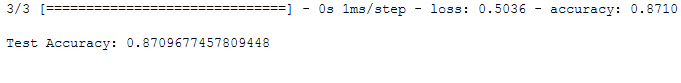
5.12 Predictions
Now it’s time for some predictions. Here I printed the first 5 results.
y_pred = model.predict(testX)
y_pred[:5]
Now we need the previously saved encoder to recode the data.
encoder_reload = pk.load(open(checkpoint_dir + '\\' + 'encoder.pkl','rb'))re_transformed_y_pred = encoder_reload.inverse_transform(y_pred)
re_transformed_y_pred[:5]
Now we can see which bird species was predicted. If you want the result to be even more beautiful, you can add the predicted values to the testX part:
testX['re_transformed_y_pred'] = re_transformed_y_pred
testX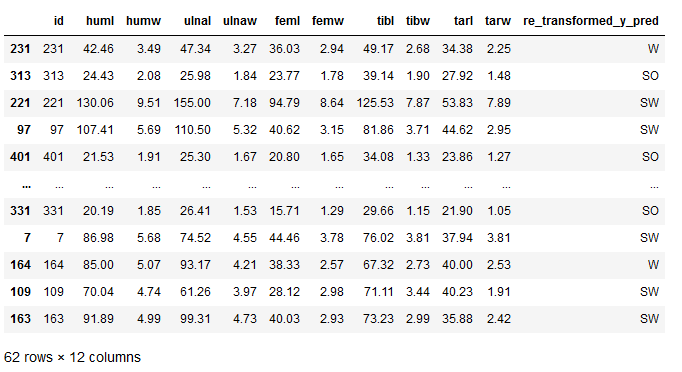
6 Prevent Overfitting
At this point I would like to remind you of the topic of overfitting. In my last post (Artificial Neural Network for binary Classification) I explained in more detail what can be done against overfitting. Here again a list with the corresponding links:
7 Conclusion
Again, as a reminder which metrics should be stored additionally when using neural networks in real life:
- Mean values of the individual predictors in order to be able to compensate for missing values later on.
- Further encoders for predictors, if categorical features are converted.
- Scaler, if these are used.
- If variables would have been excluded, a list with the final features should have been stored.
For what reason I give these recommendations can be well read in my Data Science Post. Here I have also created best practice guidelines on how to proceed with model training.
I would like to add one limitation at this point. You may have noticed it already, but the dataset was heavily imbalanced. How to deal with such problems I have explained here: Dealing with imbalanced classes
References
The content of the entire post was created using the following sources:
Chollet, F. (2018). Deep learning with Python (Vol. 361). New York: Manning.