1 Introduction
Now that we’ve gotten into the subject of ETL and I’ve shown how to call “.py files from different sources”, it’s time to write a simple but profitable ETL for data analysis.
In this article I will discuss in chapter 3 which process steps (divided into extract, transform and load) I want to do exactly and then I will show in chapter 4 how to write the previously shown steps into a .py file.
Overview of the ETL steps:

At the end I will test the created ETL. I will show this from another jupyter notebook and a fully automatic call from the command line.
For this post I use two specially created sample data sets. A copy of them is stored in my “GitHub Repo”.
2 Setup
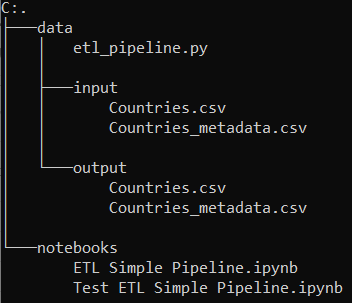
This diagram shows my usual setup. I always use a data-folder in which I insert another file ‘input’ and put my original data there. The shown output-folder is automatically created by the ETL. The etl_pipeline.py is stored in the data-folder. Furthermore I create a notebook folder from which I start the jupyter notebooks. Let’s start with the simple ETL pipeline.
3 ETL Simple Pipeline
As already announced in the introduction, I start my analysis including all ETL steps as usual in a jupyter notebook (‘ETL Simple Pipeline.ipynb’).
import pandas as pd3.1 Extract
countries = pd.read_csv('../data/input/Countries.csv')
countries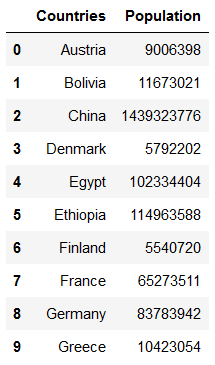
countries_metadata = pd.read_csv('../data/input/Countries_metadata.csv')
countries_metadata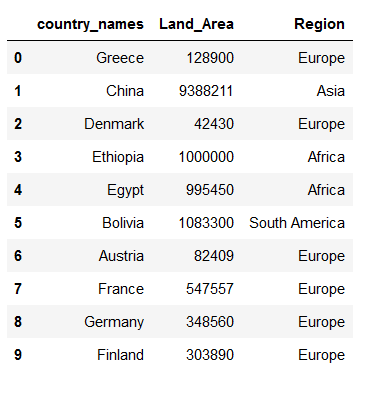
3.2 Transform
I will use only a small number of transformation steps for both datasets. These steps can be extended according to your needs and possibilities.
countries['Population'] = countries['Population']/1000
countries = countries.rename(columns={'Population':'Population_per_k'})
countries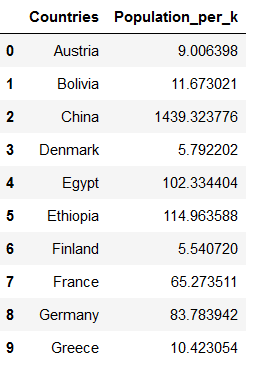
countries_metadata['Land_Area'] = countries_metadata['Land_Area']/1000
countries_metadata = countries_metadata.rename(columns={'Land_Area':'Land_Area_per_k'})
countries_metadata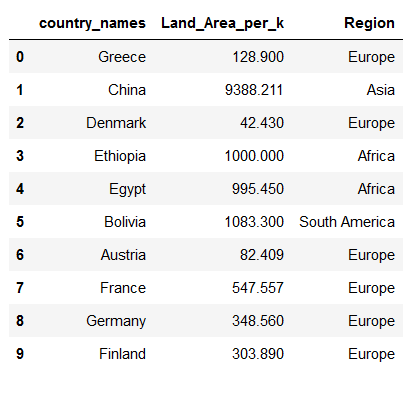
3.3 Load
countries.to_csv('../data/output/countries.csv')
countries_metadata.to_csv('../data/output/countries_metadata.csv')4 Create etl_pipeline.py
Since we either don’t want to have the complete syntax of the steps (extract, transform and load) in our analyse notebook or we want to run through these steps automatically, I write all commands in a .py file. This file will be named ‘etl_pipeline.py’ and saved under our data-folder. Important note here: all used libraries must be called again in the .py file.
import pandas as pd
import numpy as np
import os
class DataPreprocessor:
def __init__(self, path_folder = "path/to/data"):
self.path_folder = path_folder
# Path to input
self.path_input_folder = "{}/input/".format(path_folder)
self.path_input_countries = self.path_input_folder + 'Countries.csv'
self.path_input_countries_metadata = self.path_input_folder + 'Countries_metadata.csv'
# Path on which output tables are saved
self.path_output_folder = "{}/output/".format(path_folder)
self.path_output_countries = self.path_output_folder + 'Countries.csv'
self.path_output_countries_metadata = self.path_output_folder + 'Countries_metadata.csv'
# create dictionaries for read dtypes
self.read_dtypes_countries = {'Countries':'category'}
self.read_dtypes_countries_metadata = {'country_names':'category'}
# create folders for output if not existent yet
if not os.path.exists(self.path_output_folder):
os.makedirs(self.path_output_folder)
def read_data_from_raw_input(self):
print("Start:\tRead in countries Dataset")
self.countries = pd.read_csv(self.path_input_countries, dtype=self.read_dtypes_countries)
print("Finish:\tRead in countries Dataset")
print("Start:\tRead in countries_metadata Dataset")
self.countries_metadata = pd.read_csv(self.path_input_countries_metadata, dtype=self.read_dtypes_countries_metadata)
print("Finish:\tRead in countries_metadata Dataset")
def preprocess_data(self, save_preprocess_countries=True, save_preprocess_countries_metadata=True):
print("Start:\tPreprocessing countries Dataset")
self.preprocess_countries()
print("Finish:\tPreprocessing countries Dataset")
print("Start:\tPreprocessing countries_metadata Dataset")
self.preprocess_countries_metadata()
print("Finish:\tPreprocessing countries_metadata Dataset")
if save_preprocess_countries:
print("Start:\tSave countries Dataset to disc")
self.countries.to_csv(self.path_output_countries, index=False)
print("Finish:\tSave countries Dataset to disc")
if save_preprocess_countries_metadata:
print("Start:\tSave countries_metadata Dataset to disc")
self.countries_metadata.to_csv(self.path_output_countries_metadata, index=False)
print("Finish:\tSave countries_metadata Dataset to disc")
return self.countries, self.countries_metadata
def preprocess_countries(self):
self.countries['Population'] = self.countries['Population']/1000
self.countries = self.countries.rename(columns={'Population':'Population_per_k'})
def preprocess_countries_metadata(self):
self.countries_metadata['Land_Area'] = self.countries_metadata['Land_Area']/1000
self.countries_metadata = self.countries_metadata.rename(columns={'Land_Area':'Land_Area_per_k'})
def read_preprocessed_tables(self):
print("Start:\tRead in modified countries Dataset")
self.countries = pd.read_csv(self.path_output_countries, dtype=self.read_dtypes_countries)
print("Finish:\tRead in modified countries Dataset")
print("Start:\tRead in modified countries_metadata Dataset")
self.countries_metadata = pd.read_csv(self.path_output_countries_metadata, dtype=self.read_dtypes_countries_metadata)
print("Finish:\tRead in modified countries_metadata Dataset")
return self.countries, self.countries_metadata
def main():
datapreprocesssor = DataPreprocessor()
datapreprocesssor.read_data_from_raw_input()
datapreprocesssor.preprocess_data()
print('ETL has been successfully completed !!')
#if __name__ == '__main__':
# main()We have commented out the main from the ETL pipeline here with ‘#’. Of course, this syntax must not be commented out in the .py file.
Unfortunately the view is not optimal. Therefore I recommend to copy the syntax into a .py file. I prefer “Visual Studio Code” from Microsoft. But I also put the file in my “GitHub Repo” from where you can get it.
5 Test etl_pipeline.py
Now we want to test our created ETL.
5.1 from jupyter notebook
First I want to test the ETL from a notebook. For this we create and start a new notebook in the notebooks-folder with the name ‘Test ETL Simple Pipeline.ipynb’.
import sys
# Specifies the file path where the first .py file is located.
sys.path.insert(1, '../data')
import etl_pipelinedatapreprocessor = etl_pipeline.DataPreprocessor()
datapreprocessor.read_data_from_raw_input()
countries, countries_metadata = datapreprocessor.preprocess_data(save_preprocess_countries=True, save_preprocess_countries_metadata=True)
countries, countries_metadata = datapreprocessor.read_preprocessed_tables()
countries
countries_metadata
5.2 from command line
Because we have defined a main in the .py file we are able to call and execute the ETL from the command line. It does not matter which command line this is exactly. You can use Anaconda Power Shell, Git Bash, the Windwos Comman Line or anything else.
Type only the following commands in your command prompt:
cd "path/to/your/data/folder"
python etl_pipeline.pyThat’s it!
6 Conclusion
In this post I have shown a simple ETL, how to set it up and let it run through fully automated. In further publications I will present further ETL structures.